...
| Code Block | ||||
|---|---|---|---|---|
| ||||
idev -m 120180 -N 1 -A OTH21164 -r CoreNGS-Tue # or -A TRA23004 # -or- idev -m 90120 -N 1 -A OTH21164 -p development # or -A TRA23004 |
Data staging
Set ourselves up to process some yeast data data in $SCRATCH, using some of best practices for organizing our workflow.
| Code Block | ||||
|---|---|---|---|---|
| ||||
# Create a $SCRATCH area to work on data for this course, # with a sub-directory for pre-processing raw fastq files mkdir -p $SCRATCH/core_ngs/fastq_prep # Make symbolic links to the original yeast data: cd $SCRATCH/core_ngs/fastq_prep ln -s -f $CORENGS/yeast_stuff/Sample_Yeast_L005_R1.cat.fastq.gz ln -s -f $CORENGS/yeast_stuff/Sample_Yeast_L005_R2.cat.fastq.gz # or ln -s -f ~/CoreNGSsf /work/projects/BioITeam/projects/courses/Core_NGS_Tools/yeast_stuff/Sample_Yeast_L005_R1.cat.fastq.gz ln -s -f ~/CoreNGSsf /work/projects/BioITeam/projects/courses/Core_NGS_Tools/yeast_stuff/Sample_Yeast_L005_R2.cat.fastq.gz |
Illumina sequence data format (FASTQ)
GSAF gives you paired end sequencing data in two matching FASTQ format files, containing reads for each end sequenced. See where your data really is and how big it is.
| Code Block | ||||
|---|---|---|---|---|
| ||||
# the #-l oroptions lnsays -sf /work/projects/BioITeam/projects/courses/Core_NGS_Tools/yeast_stuff/Sample_Yeast_L005_R1.cat.fastq.gz ln -sf /work/projects/BioITeam/projects/courses/Core_NGS_Tools/yeast_stuff/Sample_Yeast_L005_R2.cat.fastq.gz |
Illumina sequence data format (FASTQ)
GSAF gives you paired end sequencing data in two matching FASTQ format files, containing reads for each end sequenced. See where your data really is and how big it is.
| Code Block | ||||
|---|---|---|---|---|
| ||||
# the -l options says "long listing" which shows where the link goes, # but doesn't show details of the real file ls -l #"long listing" which shows where the link goes, # but doesn't show details of the real file ls -l # the -L option says to follow the link to the real file, # -l means long listing (includes size) # -h says "human readable" (e.g. MB, GB) ls -Llh |
...
Exercise: What character in the quality score string in the FASTQ entry above represents the best base quality? Roughly what is the error probability estimated by the sequencer?
| Expand | ||
|---|---|---|
| ||
J is the best base quality score character (Q=41) It represents a probability of error of <= 1/10^4 10^4.1 or < 1/10,000 |
About compressed files
...
| Tip | ||
|---|---|---|
| ||
The asterisk character ( * ) is a pathname wildcard that matches 0 or more characters. (Read more about pathname wildcards: Pathname wildcards) |
Exercise: About how big are the compressed files? The uncompressed files? About what is the compression factor?
...
With no options, gzip compresses the file you give it in-place. Once all the content has been compressed, the original uncompressed file is removed, leaving only the compressed version (the original file name plus a .gz extension). The gunzip function works in a similar manner, except that its input is a compressed file with a .gz file and produces an uncompressed file without the .gz extension.
| Code Block | ||||
|---|---|---|---|---|
| ||||
| ||||
# if the $CORENGS environment variable is not defined
export CORENGS=/work/projects/BioITeam/projects/courses/Core_NGS_Tools
# make sure you're in your $SCRATCH/core_ngs/fastq_prep directory
cd $SCRATCH/core_ngs/fastq_prep
# Copy over a small, uncompressed fastq file
cp $CORENGS/misc/small.fq .
# check the size, then compress it in-place
ls -lh small*
gzip small.fq
# check the compressed file size
ls -lh small*
# uncompress it again
gunzip small.fq.gz
ls -lh small* |
...
| Warning |
|---|
Both gzip and gunzip are extremely I/O intensive when run on large files. While TACC has tremendous compute resources and its specialized parallel file system is great, it has its limitations. It is not difficult to overwhelm the TACC file system if you gzip or gunzip more than a few files at a time – as few as 5-6! See https://docs.tacc.utexas.edu/tutorials/managingio/ for TACC's guidelines for managing I/O. The intensity of compression/decompression operations is another reason you should compress your sequencing files once (if they aren't already) then leave them that way. |
...
One of the challenges of dealing with large data files, whether compressed or not, is finding your way around the data – finding and looking at relevant pieces of it. Except for the smallest of files, you can't open them up in a text editor because those programs read the whole file into memory, so will choke on sequencing data files! Instead we use various techniques to look at pieces of the files at a time. (Read more about commands for Displaying file contents)
The first technique is the use of pagers – we've already seen this with the more command. Review its use now on our small uncompressed file:
| Expand | |||||
|---|---|---|---|---|---|
| |||||
|
...
If you start less with the -N option, it will display line numbers Numbers. Using the -I options allows pattern searches to be case-Insensitive.
Exercise: What line of small.fq contains the read name with grid coordinates 2316:10009:100563?
...
| Expand | |||||
|---|---|---|---|---|---|
| |||||
|
...
| Code Block | ||||
|---|---|---|---|---|
| ||||
# shows 1st 10 lines
head small.fq
# shows 1st 100 lines -- might want to pipe this to more to see a bit at a time
head -100 small.fq | more |
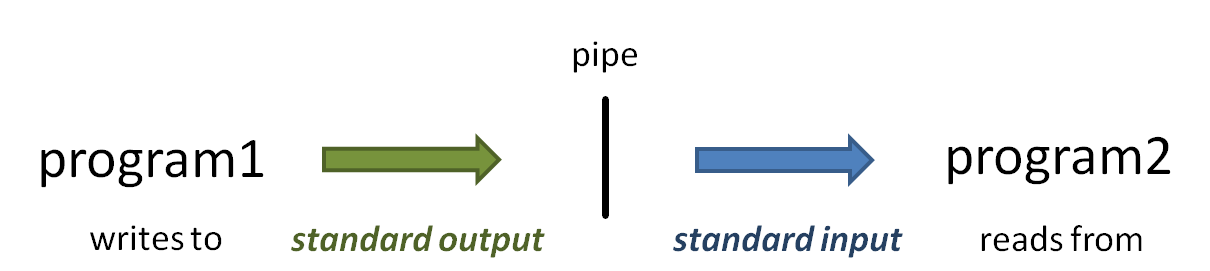
The vertical bar ( | ) above is the pipe operator, which connects one program's standard output to the next program's standard input. (Read more about Piping)
time
head -100 small.fq | more |
So what if you want to see line numbers on your head or tail output? Neither command seems to have an option to do this.
| Expand | ||
|---|---|---|
| ||
cat --help | more |
| Expand | |||||
|---|---|---|---|---|---|
| |||||
|
piping
So what is that vertical bar ( | ) all about? It is the pipe operator!
The pipe operator ( | ) connects one program's standard output to the next program's standard input. The power of the Linux command line is due in no small part to the power of piping. Read more about piping here: Piping. And read more about standard (Read more about Piping and Standard Unix I/O streams here: Standard Streams in Unix/Linux.)
- When you execute the head -100 small.fq | more command, head starts writing lines of the small.fq file to standard output.
- Because of the pipe, the output does not go to the terminal Terminal, but is connected to the standard input of the more command.
- Instead of reading lines from a file you specify as a command-line argument, more obtains its input from standard inputfrom standard input.
- The more command writes a page of text to standard output, which is displayed on the Terminal.
| Tip | ||
|---|---|---|
| ||
Most Linux commands are designed to accept input from standard input in addition to (or instead of) command line arguments so that data can be piped in. Many bioinformatics programs also allow data to be piped in. Typically Often they will require you provide a special argument, such as stdin or -, to tell the program data is coming from standard input instead of a file. |
tail
The yang to head's ying is tail, which by default it displays the last 10 lines of its data, and also uses the -NNN syntax to show the last NNN lines. (Note that with very large files it may take a while for tail to start producing output because it has to read through the file sequentially to get to the end.)
But what's really cool about tail is its -n +NNNNN syntax. This displays all the lines starting at line NNN NN. Note this syntax: the -n option switch follows by a plus sign ( + ) in front of a number – the plus sign is what says "starting at this line"! Try these examples:
| Expand | |||||
|---|---|---|---|---|---|
| |||||
|
...
| Code Block | ||||
|---|---|---|---|---|
| ||||
# shows the last 10 lines tail small.fq # shows the last 100 lines -- might want to pipe this to more to see a bit at a time tail -100 small.fq | more # shows all the lines starting at line 900 -- better pipe it to a pager! # cat -n adds line numbers to its output so we can see where we are in the file cat -n small.fq | tail -n +900 | more # shows 15 lines starting at line 900 because we pipe to head -15 tail -n +900 small.fq | head -15 |
Read more about head and tail in Displaying file contents.
zcat and gunzip -c tricks
Ok, now you know how to navigate an un-compressed file using head and tail, more or less. But what if your FASTQ file has been compressed by gzip? You don't want to un-compress the file, remember?
...
| Expand | |||||
|---|---|---|---|---|---|
| |||||
|
...
| Code Block | ||||
|---|---|---|---|---|
| ||||
zcat Sample_Yeast_L005_R1.cat.fastq.gz | more
zcat Sample_Yeast_L005_R1.cat.fastq.gz | less -N
zcat Sample_Yeast_L005_R1.cat.fastq.gz | head
zcat Sample_Yeast_L005_R1.cat.fastq.gz | tail
zcat Sample_Yeast_L005_R1.cat.fastq.gz | tail -n +901 | head -8
# include original line numbers
zcat Sample_Yeast_L005_R1.cat.fastq.gz | cat -n | tail -n +901 | head -8 |
...
| Tip |
|---|
There will be times when you forget to pipe your large zcat or gunzip -c output somewhere – somewhere – even the experienced among us still make this mistake! This leads to pages and pages of data spewing across your terminal Terminal. If you're lucky you can kill the output with Ctrl-c. But if that doesn't work (and often it doesn't) just close your Terminal window. This terminates the process on the server (like hanging up the phone), then you just can log back in. |
...
One of the first thing to check is that your FASTQ files are the same length, and that length is evenly divisible by 4. The wc command (word countword count) using the -l switch to tell it to count lines, not words, is perfect for this. It's so handy that you'll end up using wc -l a lot to count things. It's especially powerful when used with filename wild carding.
| Expand | |||||
|---|---|---|---|---|---|
| |||||
|
...
| Code Block | ||||
|---|---|---|---|---|
| ||||
echo $((2368720 / 4)) |
Here's another trick: backticks backtick evaluation. When you enclose a command expression in backtick quotes ( ` ) the enclosed expression is evaluated and its standard output substituted into the string. (Read more about Quoting in the shell).
Here's how you would combine this math expression with zcat line counting on your file using the magic of backtick evaluation. Notice that the wc -l expression is what is reading from standard input.
| Expand | |||||
|---|---|---|---|---|---|
| |||||
|
...
| Warning | ||
|---|---|---|
| ||
Note that arithmetic in the bash shell is integer valued only, so don't use it for anything that requires decimal places!decimal places! (Read more about Arithemetic in bash) |
A better way to do math
Well, doing math in bash is pretty awful – there has to be something better. There is! It's called awk, which is a powerful scripting language that is easily invoked from the command line.
In the code below we pipe the output from wc -l (number of lines in the FASTQ file) to awk, which executes its body (the statements between the curly braces ( { } ) for each line of input. Here the input is just one line, with one field – the line count. The awk body just divides the 1st input field ($1) by 4 and writes the result to standard output. (Read more about awk in Advanced Some Linux commands: awk)
| Expand | |||||
|---|---|---|---|---|---|
| |||||
|
...
| Code Block | ||||
|---|---|---|---|---|
| ||||
cd $SCRATCH/core_ngs/fastq_prep
zcat Sample_Yeast_L005_R1.cat.fastq.gz | wc -l | awk '{print $1 / 4}'
|
Processing multiple compressed files
You've probably figured out by now that you can't easily use filename wildcarding along with zcat and piping to process multiple files. For this, you need to code a for loop in bash. Fortunately, this is pretty easy. Try this:
| Code Block | ||||
|---|---|---|---|---|
| ||||
for fname in *.gz; do
echo "Processing $fname"
echo "..$fname has `zcat $fname | wc -l | awk '{print $1 / 4}'` sequences"
done |
...
Each time through the for loop, the next item in the evaluation expression (here *.gz) is assigned to the formal argument (here fname). Note that items in the evaluation expression are separated by spaces – not Tabs.
...
The bash shell lets you put multiple commands on one line if they are each separated by a semicolon ( ; ). So in the above for loop, you can see that bash considers the do keyword to start a separate command. Two alternate ways of writing the loop are:
| Code Block | ||
|---|---|---|
| ||
# One line for each clause, no semicolons
for <variable name> in <expression>
do
<something>; <something else>
done |
...
| language | bash |
|---|
...
Note that $1 means something different in awk – the 1st whitespace-delimited input field – than it does in bash, where it represents the 1st argument to a script or function (technically, the 1 environment variable). This is an example of where a metacharacter- the dollar sign ( $ ) here – has a different meaning for two different programs. (Read more about Literal characters and metacharacters)
The bash shell treats dollar sign ( $ ) as an evaluation operator, so will normally attempt to evaluate the environment variable name following the $ and substitute its value in the output (e.g. echo $SCRATCH). But we don't want that evaluation to be applied to the {print $1 / 4} script argument passed to awk; instead we want awk to see the literal string {print $1 / 4} as its script. To achieve this result we surround the script argument with single quotes ( ' ' ), which tells the shell to treat everything enclosed by the quotes as literal text, and not perform any metacharacter processing. (Read more about Quoting in the shell)
Processing multiple compressed files
You've probably figured out by now that you can't easily use filename wildcarding along with zcat and piping to process multiple files. For this, you need to code a for loop in bash. Fortunately, this is pretty easy. Try this:
| Code Block | ||||
|---|---|---|---|---|
| ||||
cd $SCRATCH/core_ngs/fastq_prep
for fname in *.gz; do
echo "Processing $fname"
echo "..$fname has `zcat $fname | wc -l | awk '{print $1 / 4}'` sequences"
done |
Each time through the for loop, the next item in the argument list (here the files matching the wildcard glob *.gz) is assigned to the for loop's formal argument (here the variable fname). The actual filename is then referenced as$fname inside the loop. (Read more about Bash control flow)